omni_synthregressionmask#
Overview#
omni_synthregressionmask is a utility designed to assist with generating the weight images used by omni_synthtarget when estimating synthetic EPI images. omni_synthregressionmask is used in the omni preprocessing scripts, omni_synthpipeline and omni_synthpreproc. The goal of a good weight image is to reduce the contribution of very noisy or distorted areas when estimating a synthetic image. Properly created, a good weight mask can significantly improve the accuracy of the contrast properties of a synthetic image. Good synthetic EPI contrast accuracy is key to successfully performing field map-less distortion correction. Understanding how omni_synthregressionmask works may be helpful for users who are incorporating field map-less distortion correction into a pre-exisiting MRI preprocessing pipeline or dealing with tricky registration cases where default parameters are not performing optimally.
Note
Correctly estimating the voxel intensity mapping needed to create a synthetic EPI image is very difficult when source and and target images are distorted differently. This is because in highly distorted areas corresponding voxels in source and target images may no longer represent the same portion of the imaged object. When this occurs, voxels in the mismatched portions of the target and source images can no longer be accurately modelled by the true source(s)-to-target intensity mapping function. Such areas are effectively noise and tend to reduce the contrast of the resulting synthetic image.
Also, it is often desirable to bias the estimate of the synthetic EPI image so that it favors the accurate representation of some particular region of the target image. For instance, when creating a synthetic EPI image intended for field map-less distortion correcton, you may want to ensure that the contrast properies of the part of the synthetic image containing the brain is the most accurate, and permit relatively lower contrast accuracy in the region on the synthetic image containing the neck and eyeballs. This can be accomplished by providing omni_synthtarget with an appropriate weight image, such as the one produced by omni_synthregressionmask.
Example commands#
# Example 1:
omni_synthregressionmask -e ~/data/EPIData.nii.gz -m ~/data/AnatomicalBETMask.nii.gz -a Anat2EPI.1D -o ~/data/RegressionMask.nii.gz
# Example 2:
omni_synthregressionmask -e ~/data/EPIData.nii.gz -m ~/data/AnatomicalBETMask.nii.gz -a Anat2EPI.1D -o ~/data/RegressionMask.nii.gz -f Anat2EPINonLinearWarp.nii.gz -s 6
Illustration#
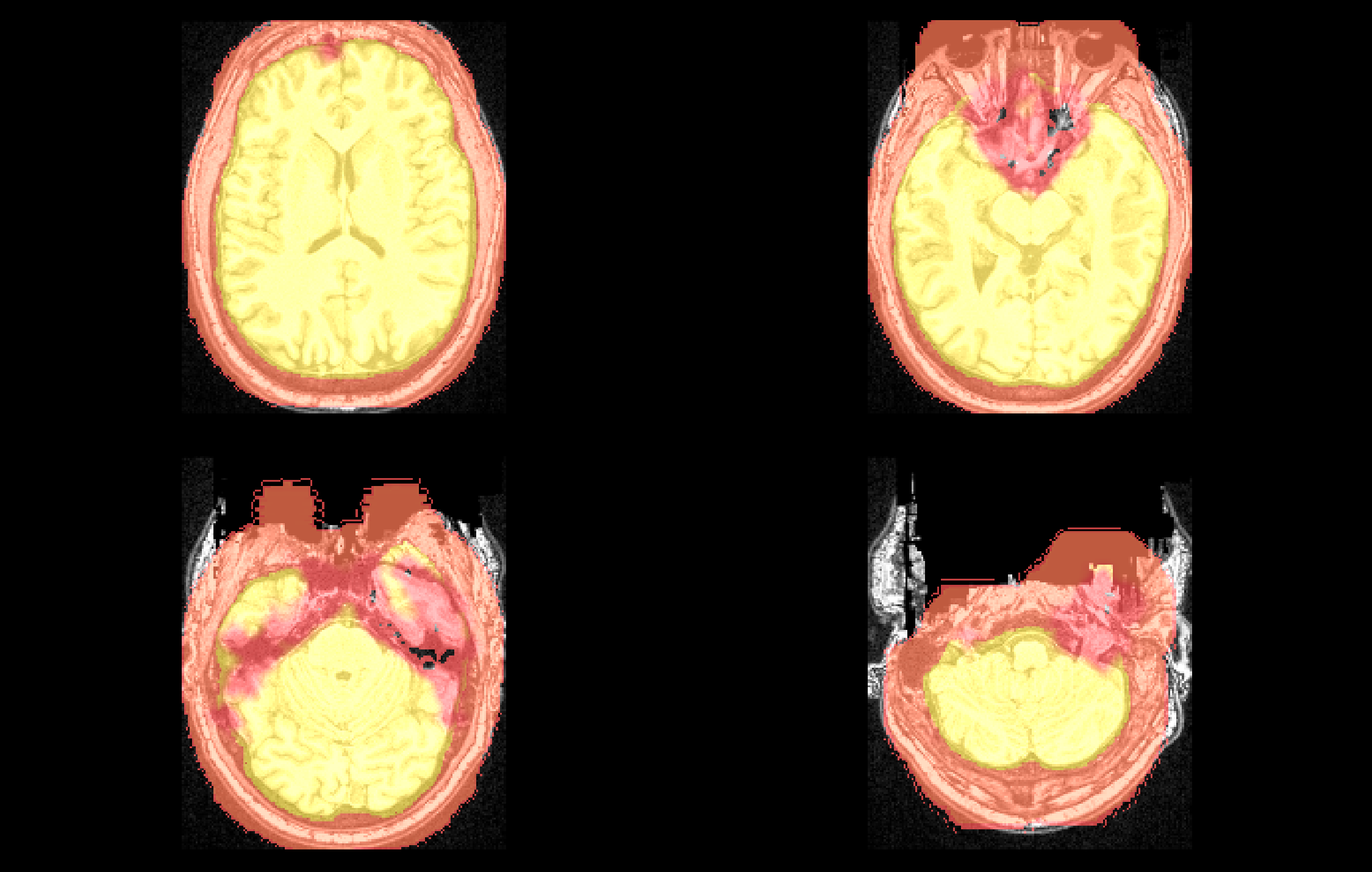
Example weight mask produced by omni_synthregressionmask. A weight mask emphasizing the contribution of brain voxels. Reduced weighting is depicted in red, increased weighting is depicted in yellow. omni_synthregressionmask produces weight masks that reduce the contribution of noise regions (internally detected with omni_synthnoisemask) and increase the contribution of brain voxels relative to immediately the surrounding skull.#
Arguments#
Required arguments#
- dil_anat_bet_mask
The file name of a dilated brain only mask aligned to an anatomical (typically T1w or T2w) image. Only voxels within this mask will contribute anything at all to the final weighting image. Generally this mask should represent the entire brain as well as a portion of the skull (and perhaps a little beyond that too). This is meant to exclude voxels far outside of the head that we do not wish to consider when estimating a synthetic image.
- anat_bet_mask
The file name of a binary brain only mask aligned to an anatomical (typically T1w or T2w) image. Voxels within this mask are given the greatest (2x) significance in the final weighting image.
- epi
The file name of an EPI data set (e.g., task, rest or diffusion weighted). This data set must contain multiple frames. Any desired frame-wise alignment and slice timing correction should already be performed.
- anat_to_epi_affine
Path to a file representing an affine transformation that aligns the anatomical image to the EPI data set. This text file must represent the affine transformation in AFNI format (i.e., suitable for use with 3dAllineate).
- output_path
The directory where output files should be stored.
Optional arguments#
- -w WARP, --warp WARP
A non-linear transformation that will warp an affine-aligned anatomical image into proper registration with the input EPI data set. The inverse of this warp is applied to the weighting image (when it is aligned to the EPI data set) before it is transformed back into the anatomical image space and saved out. Including this optional warp is useful in situations where one is iteratively correcting distortion and updating the synthetic image during each step (as in
omni_synthunwarpandomni_synthpreproc). In that case, regions that were initially determined to be noise and were down weighted, may have shifted by some amount as a result of earlier corrections. Including the previous corrective warp as input ensures that weight mask stays in register with current distortion correction iteration.
- -d NOISE_MASK_DILATION_SIZE, --noise_mask_dilation_size NOISE_MASK_DILATION_SIZE (default: 2)
NOISE_MASK_DILATION_SIZEis an integer value that determines how many voxels to dilate the noise mask before writing it to a file. This option can be used to create more “conservative” noise masks that label voxels as suspect by simply by virtue of their proximity to real noise voxels. It is left to the user to contemplate the morality of this guilt-by-association approach.
- -i NOISE_MASK_ITERATIONS, --noise_mask_iterations NOISE_MASK_ITERATIONS (default: 12)
NOISE_MASK_ITERATIONSis an integer that sets the number of refining iterations used in the the noise mask generating LDA procedure. The initial ‘guess’ for what voxels contain valid signal is typically a binary brain only mask derived from the subjects anatomical images and then aligned to the EPI image. This initial guess is then refined using an iterative temporal linear discriminant analysis approach. In our our experiments a stable noise mask is typically achieved in 8-10 iterations. For good measure, we set the default value just a little higher than this. The appropriate number of iterations may vary depending on the data type (resting state vs diffusion type EPI data) as well as the number of samples (frames) present in the data. If you want to make every second count, you can probably reduce the default without causing too much havoc. If the synthetic images produced by omni_synthpreproc do not appear sufficiently accurate for your tastes, we advise that you look closely at the noise mask volumes that are being estimated for your data set to make sure that voxels that are being labelled as noise are sensible. Do this by loading up the EPI data in your favorite viewer and using the estimated noise mask images as an overlay.
- -s NOISE_MASK_SIGMA, --noise_mask_sigma NOISE_MASK_SIGMA (default: 2)
NOISE_MASK_SIGMAdefines the standard deviation of a Gaussian kernel used to smooth the binary noise mask before writing it to a file. This option is helpful for creating weight volumes rather than simply binary labels. Gaussian smoothing is performed after any noise mask dilation
- -v, --version
Show program’s version and exit.
- -h, --help
show this help message and exit