omni_synthnoisemask#
Overview#
omni_synthnoisemask is a utility that is used to automatically detect and
label regions of an EPI data set that are likely to be affected by unacceptable
levels of signal drop out and noise. The binary masks produced by this utility
are useful for a variety of purposes, however it was designed initially to
improve the quality of synthetic EPI images used for field map-less distortion
correction by reducing the contribution of highly distorted areas when
estimating the voxel intensity mapping between source images (e.g., undistorted
T1w and T2w images) and a target image (e.g., a low resolution distorted EPI).
Note
omni_synthnoisemask creates noise mask using the following procedure: An
affine aligned anatomical brain mask is used as an initial signal and noise
labeling. All voxels in this this mask labeled with ones are initially
considered to be “signal”; all voxels labeled with zeros are considered to
be “noise”. A between-class linear discriminant analysis (LDA) is run on the
raw (un-centered and un-scaled) EPI time series for all of the voxels (this
is why it is important that frame-wise alignement and slice time correction
be performed first). Each voxel’s time series is then projected onto the
principal discriminant vector. The procedure is repeated, this time using
the centered and normalized voxel time series. The centered and normalized
time voxel time series are projected onto the new principal discriminant
vector. Each voxel at this point is associated with two projection values. A
third value is created for each voxel equal to the product of the two LDA
projection values. A second level LDA is then performed using these three
values and each voxel’s triplet of values is projected onto the resulting
principal discriminant vector. Otsu’s method is then used to classify each
voxel as signal or noise depending on its projection onto this vector and
produce an updated label for each voxel. This process is then repeated.
Sufficiently iterated the signal and noise labels produced by this method
converge and stabilize.
Example commands#
# Example 1:
omni_synthnoisemask ~/data/EPIData.nii.gz ~/data/AlignedAnatomicalBETMask.nii.gz ~/data/EPINoiseMask.nii.gz
# Example 2:
omni_synthnoisemask ~/data/EPIData.nii.gz ~/data/AlignedAnatomicalBETMask.nii.gz ~/data/EPINoiseMask.nii.gz -d 3 -k 8
Illustration#
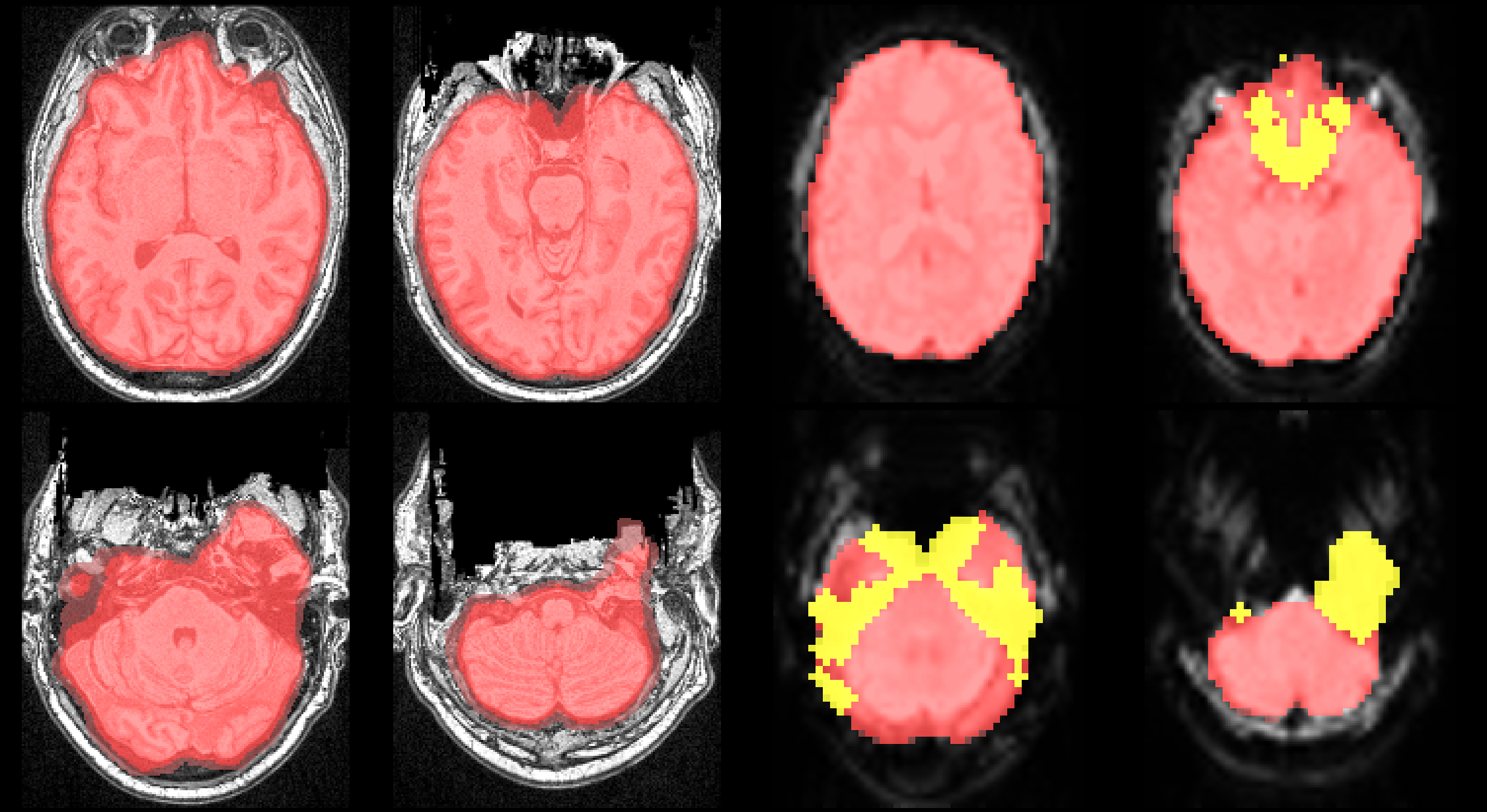
Initial anatomical brain mask and resulting noise mask Left: the anatomically derived brain mask (red) is displayed as an overlay on the subject’s T1w image. Right: the brain mask pre-aligned to the EPI data data set. The pre-aligned mask indicates the relevant region of in which to detect noise and is an initial labeling of potentially valid voxels. The resulting binary noise mask (yellow) produced after 12 iterations.#
Arguments#
Required arguments#
- epi
The file name of an EPI data set (e.g., task, rest or diffusion weighted). This data set must contain multiple frames. Any desired frame-wise alignment and slice timing correction should already be performed.
- anat_bet_mask_epispace
The file name of a binary brain mask defined by the subjects undistorted anatomical images (e.g., T1w or T2w or some composite of them) that has been affine aligned to the EPI data set referred to by the first,
epi, argument.It provides a very rough initial labeling of EPI voxels that are likely to be valid and relatively noiseless.
It labels the region of the EPI image in which we are interested in detecting noise. That is, the primary use case for
omni_synthnoisemaskis to detect noisy voxels in the area where the brain should be.
- output
Path to output file containing the estimated noise mask.
Optional arguments#
- -i NOISE_MASK_ITERATIONS, --noise_mask_iterations NOISE_MASK_ITERATIONS (default: 12)
NOISE_MASK_ITERATIONSis an integer that sets the number of refining iterations used in the the noise mask generating LDA procedure. The initial ‘guess’ for what voxels contain valid signal is typically a binary brain only mask derived from the subjects anatomical images and then aligned to the EPI image. This initial guess is then refined using an iterative temporal linear discriminant analysis approach. In our our experiments a stable noise mask is typically achieved in 8-10 iterations. For good measure, we set the default value just a little higher than this. The appropriate number of iterations may vary depending on the data type (resting state vs diffusion type EPI data) as well as the number of samples (frames) present in the data. If you want to make every second count, you can probably reduce the default without causing too much havoc. If the synthetic images produced by omni_synthpreproc do not appear sufficiently accurate for your tastes, we advise that you look closely at the noise mask volumes that are being estimated for your data set to make sure that voxels that are being labelled as noise are sensible. Do this by loading up the EPI data in your favorite viewer and using the estimated noise mask images as an overlay.
- -d NOISE_MASK_DILATION_SIZE, --noise_mask_dilation_size NOISE_MASK_DILATION_SIZE (default: 2)
NOISE_MASK_DILATION_SIZEis an integer value that determines how many voxels to dilate the noise mask before writing it to a file. This option can be used to create more “conservative” noise masks that label voxels as suspect by simply by virtue of their proximity to real noise voxels. It is left to the user to contemplate the morality of this guilt-by-association approach.
- -k NOISE_MASK_SIGMA, --noise_mask_sigma NOISE_MASK_SIGMA (default: 2)
NOISE_MASK_SIGMAdefines the standard deviation of a Gaussian kernel used to smooth the binary noise mask before writing it to a file. This option is helpful for creating weight volumes rather than simply binary labels. Gaussian smoothing is performed after any noise mask dilation
- -v, --version
Show program’s version and exit.
- -h, --help
show this help message and exit